
Interpreting DNA RESULTS
>
PARENT VERIFICATION RESULTS
Parent verification is reported back to the member with the following;
Qualified - If qualified is shown next to a Sire or Dam it is indicating the the lab has confirmed that this is the parent
Excluded - If shown next to a Sire or Dam it is indicating the the lab has confirmed that this is not the parent
If you have tested potential parents and the lab results qualify a different parent to the one listed on the database the office will automatically process the pedigree change.
> GENERAL
N – Non-Carrier DNA analysis is consistent with the subject NOT carrying the mutation tested
C – Carrier DNA analysis is consistent with the subject CARRYING the mutation tested
A – Affected DNA analysis is consistent with the subject carrying two copies of the mutation tested
NR – No Result
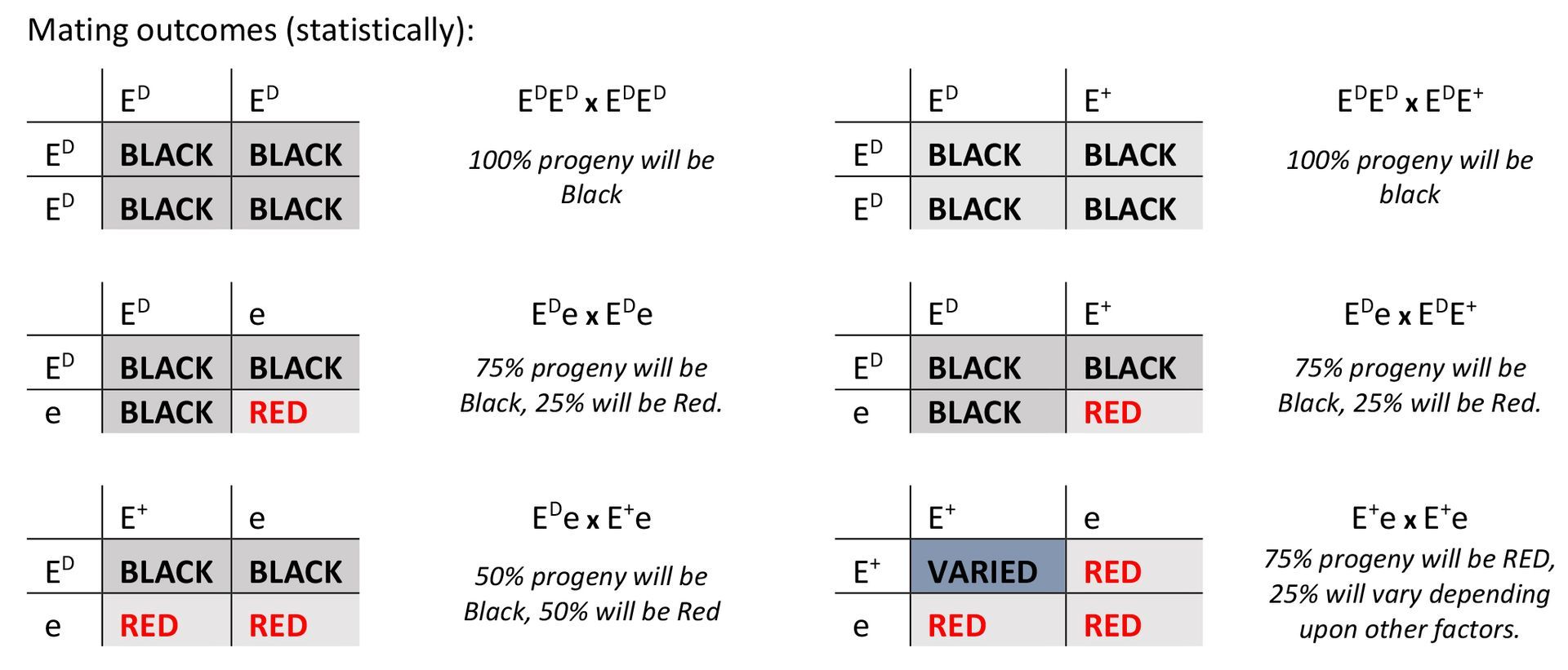
> COAT COLOUR
Coat colour testing is reported back to the member with the following
- ED Homozygous dominant black
- ED/e dominant black/recessive red
- ED/E+ dominant black/wildtype
- E+/e wildtype/recessive red
- E+/E Homozygous wildtype
- e/e Homozygous recessive red
Several genes are involved in the process of creating the complex coat colours and patterns found in domestic cattle.
One of these is the Melanocortin 1 Receptor (MC1R) gene, also called Extension, that controls the production of black (eumelanin) and red (phaeomelanin) pigments. The three alleles (forms) of this gene are:
- Dominant black (ED) - Dominant to the other two alleles and animals with this allele are jet black (solid or spotted).
- Recessive red (e) - Two copies of this (e) allele will result in red colour.
- Wild type (E+) – the ancestral/’wild-type’ allele "E+" is neutral, producing a black coat in the presence of "ED",a red coat in the presence of "e" and a variety of colours in E+/E+ animals, where other genes also influence the pigments produced.
| Genotype | Description | Colour |
|---|---|---|
| ED/ED | Homozygous Dominant Black | Black |
| ED/E+ | Dominant Black/Wildtype | Black |
| ED/e | Dominant Black//Recessive Red | Black |
| E+/e | Wildtype/Recessive Red | Red |
| E+/E+ | Homozygous Wildtype | Varied* |
| e/e | Homozygous Recessive Red | Red |
NOTE: Other coat colour genes act as modifiers of these base colours adding white spotting patterns, reorganizing the distribution of red and black pigments (Brindle and Agouti) or diluting the pigments (Dun, Charolais dilution and Simmental dilution). For example, the PMEL17_delTTC allele causes dilution coat colours such as dun, silver dun, yellow, and cream based on an interaction with the MC1R gene. This dilution factor is common in multiple breeds including Charolais, Hereford, Highland, Galloway, and Simmental.
> POLL RESULTS
Poll testing is reported back to the member with the following:
- PcPc, PcPf and PfPf Homozygous poll
- Hpc and Hpf Heterozygous poll
- HH Horned
- NR Failed test or due to unresolved haplotype (combination of HornPoll markers fail to produce valid result)
> TENDERNESS
Increase in 'tenderness' is associated with favourable alleles seen within the selected marker panel. In this report, the combined genotype results have been scored between 1 to 10, where 10 is the most favourable number of alleles present.
To discuss DNA results further please contact the ABCA office on 02 5775 9900 or email office@brangus.com.au
DISCLAIMER
QUICK LINKS
THE BRANGUS ADVANTAGE BECOME A MEMBER TODAY AUSTRALIAN BRANGUS MAGAZINES WHAT IS BREEDPLAN? HOW TO SAMPLE DNA
© Copyright 2024 All Rights Reserved Australian Brangus Cattle Association Ltd















